Online Platform for Clinical and Molecular data Retrieval
Introduction
This post is about describing a technologies stack used in a project aimed to create an online platform for clinical and molecular data retrieval. The online platform also includes a functionality to analyse molecular data stored in the cloud. The context of the project is in a biomedicine company that wants to provide to clients and to potential clients with a tool to explore the available data.
Functionality requirements
The objective is to provide to client with a very resopnsive interface that, to begin with, will show an overview of the quantity and types of data that we have.
For this, an interactive dashboard with the basic data measurements is set as the landing page.
In our case, the most important concept in a search is the disease. This first step of choosing the disease is enabled by a drop down list. From there, the client can select with a faceted search the group of donors to get information from. The faceted search includes both, clinical variables of the donors, (“donors” tab), and kind of files, (“files” tab), to select.
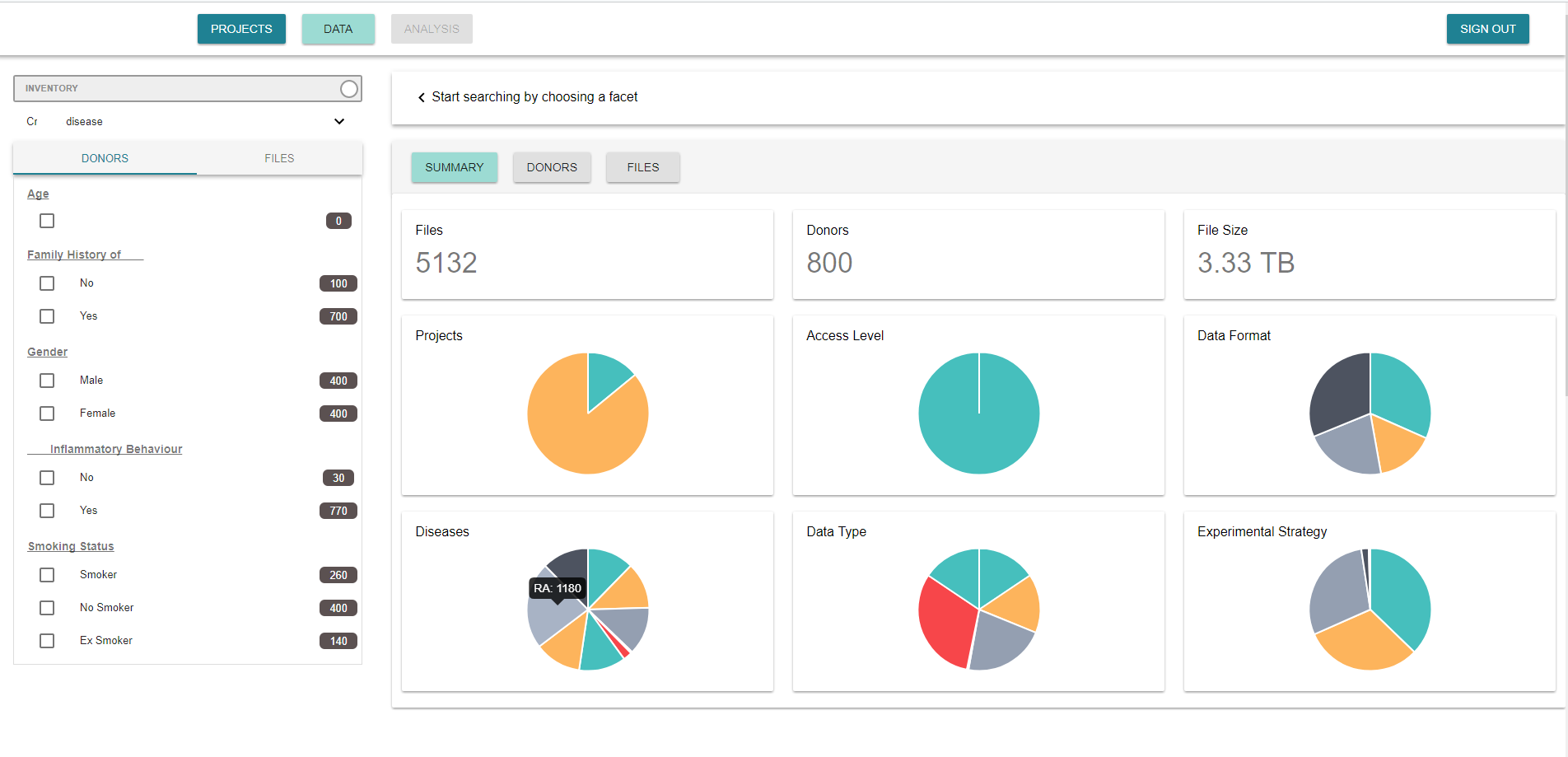
Additionally, there is a switch that allows to select between two modes in the platform. Whereas in the inventory mode the client can see basic information of the date, in the exploration mode, the client can fully access to data, from which they already have purchased the rights to us.
Technologies Stack
Data Inventory
The first technologies stack presented here includes everthing required to enable the client to see what data do we have.
With the platform feature requirements, the stack is started to be built from its bottom, finding out the more convenient technologies to store the data. On the one hand the clinical data, which are variables from donors, such as the gender, age, etc. and on the other hand the molecular data, which is in the order of GB or TB for each donor. In this case, GCP storage services have been used.
With this, it appears the need of relating the clinical data with the molecular data. For example: for each donor, what are their clinical variables and their molecular data files? To solve this problem, a metadata file is asigned to each molecular file. This metadata file contains information related to the molecular data file such as the donor identification (previously de-identified), the assigned project, data processing methodolody, etc. In the case, of the clinical data, those data are already in SQL format, which can be easily imported to GCP SQL.
Having metadata in the form of files and clinical data in SQL tables leads to the question of how to make queries to both data at the same time in an efficient manner. Because of the data characterization could be modified in a near future and that each donor could have very different characterization in terms of data variables, the decision has been to use a hibrid database model. In this case OrientDB has been used because of its high functional and technological flexibility.
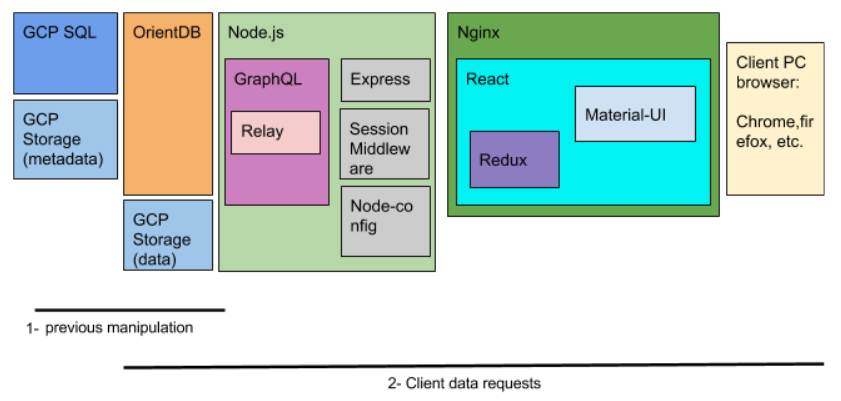
As the above image shows, the technologies explained until now, correspond to a previous manipulation of the data in our side. The second line, indicates the pieces that come into action when the client requests data. To access the databse in the backdrop, a Node.js server with Express framework has been used. To make the queries to the OrientDB more straighforward in terms of development, GraphQL query language is used. In GraphQL, the developer has to define the data model that exists in the underlying database.
On the side of the application, React in conjunction with Redux state machine have been used. This allows to refresh in the page only the components that have to undergone some change because of an action of the client. The application is placed in a Nginx server, which allows the application to scale in a smarter way than using an Apache server.
Data Analtytics
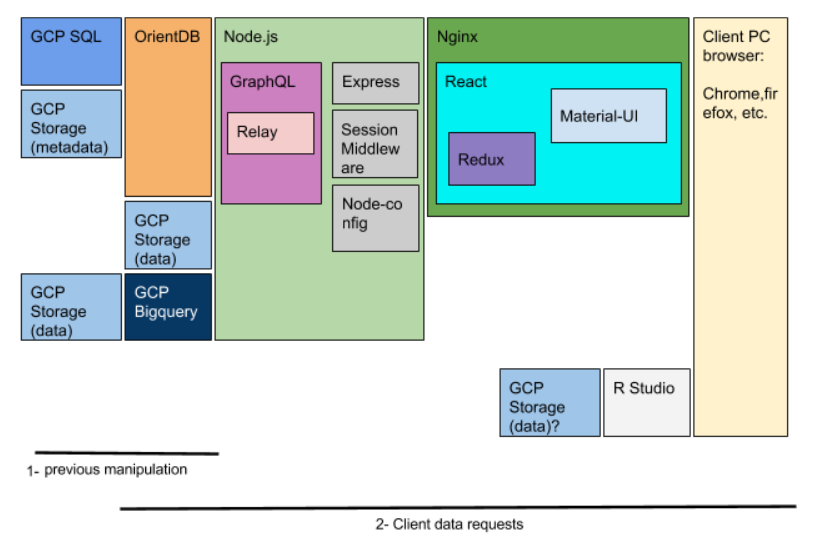
To build the data analytics funcionality, big data technologies have been used because the involved data are of the order of TB. In this case, the decision has been to use GCP Bigquery, therefore data are needed to be ingested in the GCP Bigquery. This allows to develop ad-hoc analyses on the side of the front-end. Beyond doing these ad-hoc analyses, there is also a need to allow the client to make their own analyses. Although this functionality highly requires securing the enviroment, a proposal is to use R-studio server via web. This part is not explained in this post because protecting the R access by using a remote virtual desktop environment has appeared to be more convenient and secure, though this is part of another project.
DevOps
This section explains how the whole stack is interconnected, what procedures are needed to keep the system running as well as the underlying hardware platform that supports the stack.
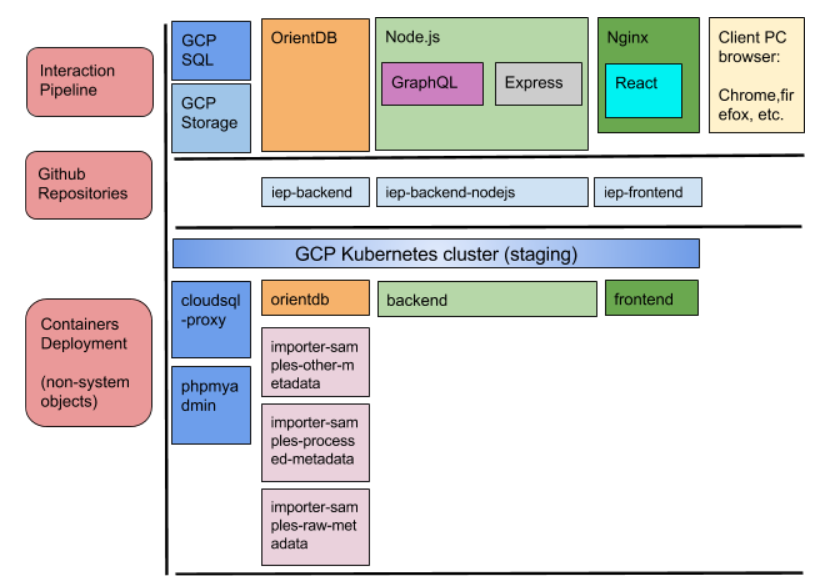
As the figure below shows, the code is gathered in different packages that are managed using GitHub repositories. In this case, the repositories are connected to GCP through source repositories GCP service. Thanks to this, each time a developer pushes a change in the code, the system running in GCP gets updated by building what is new in the repositories.
The used infrastructure technology are Docker containers managed with Kubernetes on top of GCP VM instances. This gives modularity to the stack and very light and responsive VM instances, as the used operating systems only include what is needed to run each piece of the stack.
The OrientDB container also includes scripts that are run when there are updates in the SQL database or when more molecular data are uploaded to the GCP buckets. Those scripts update the OrientDB graph so all the relations are set correctly making possible that the client can get the right results when using the faceted search.
Caveats
This stack has a node.js step between the DB layer and the application server layer. This adds potential to use GraphQL to query OrientDB. Howerver, at the same time it represents and additional step that adds an extra of latency. In this case, the client petitions delayed almost one second. The problem arises when the client concatenates several requests when selecting and desselecting facets in the menu. If the petitions are just queued, the latency starts to be unacceptable for an online platform, reaching waiting times of 4 to 5 seconds. To solve this situation, the petitions that haven’t been delivered when a new one comes are discarted, as the new petition always contains the whole set of facets to query to OrientDB.





